The text below outlines the research projects I pursued as a PhD student and postdoctoral fellow. Since 2020, I have shifted my focus towards supporing high-performance computing projects at the Marlyand Advanced Research Computing Center (MARCC) and more recent work on computational cardiology with Natalia Trayanova and the Alliance for Cardiovascular Diagnostic and Treatment Innovation. My HPC work has largely been customized to clusters at MARCC, however I co-authored a tool for integrating Singularity containers with Lmod modules called Community Collections. My contribution to Professor Trayanova’s personalized computational cardiology work is ongoing.
computational biophysics research
I’ve had the pleasure of participating in several distinct research projects which tend to orbit the question of how biological systems work.
How do very small objects (molecules), affect larger, more complex biological systems (from organelles to organisms) in measurable ways?
My research is organized by topic areas below – I’ve tried to cast a wide net. The capstone of my Ph.D. thesis is a formal method for measuring protein-induced membrane curvature. I am now applying this work to an ongoing study of the proteins that generate filopodia. In addition to these coarse-grained simulations, I have used atomistic methods to study the chemistry of phosphoinositides. I have also measured the flexibility of intracellular adhesion molecules to inform a model for particles designed to bind certain kinds of cells. As an undergraduate I developed a model for understanding the spread of malaria in a host population according to how the virus replicates in the bloodstream.
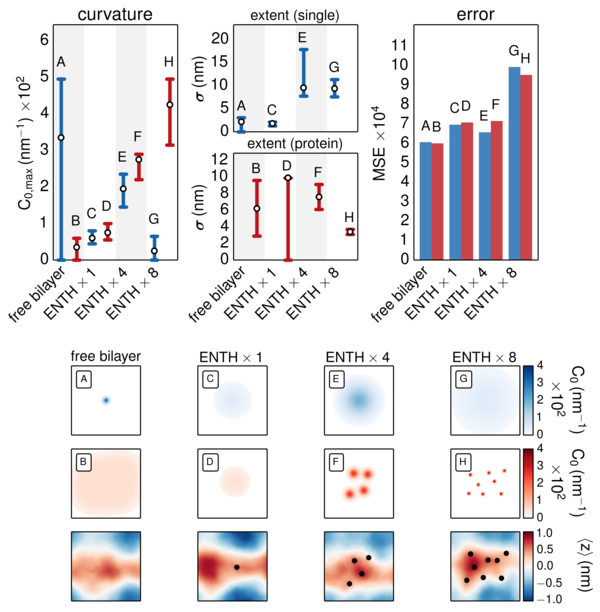
Measuring curvature induced by part of the protein epsin in coarse-grained molecular dynamics simulations. The red fuzzy pin-pricks (B-H) represent the induced deformations.
proteins focus curvature
There is much interest in understanding how proteins self-assemble into complexes which bend cell membranes during intracellular trafficking events. In order to predict the conditions necessary to trigger intracellular trafficking events, one must be able to quantify both curvature sensing and generation by protein complexes in terms of their chemical identity, concentration, and arrangement. My advisor Ravi Radhakrishnan and I have developed a method for calculating protein-induced membrane curvature using coarse-grained molecular dynamics simulations. In particular, we distinguish thermal undulations from induced curvature according to the coupling between them expressed by the Helfrich Hamiltonian. This method can be used to predict the cooperative concentration-curvature relationship necessary to predict membrane morphology change. Interestingly, we find that these long-mode undulations facilitate interactions that are much larger than the proteins themselves. The resulting curvature-undulation coupling may provide a physical mechanism for long-ranged recruitment of curvature sensing proteins which generate and stabilize mature membrane morphologies in cells. This method is general to many membrane-remodeling protein systems and may help to inform more detailed cell signaling models. This research is currently in press.

molecular mechanism of exocyst-mediated filapodia
In a collaboration between several research groups on campus, I have used molecular modeling to predict the formation of filapodia, cellular protrusions. We have traced the formation of specific kinds of protrusions to the exocyst complex. A key member of the complex (exo70) is responsible for pinching the cell membrane outwards to create space absent the force of the cytoskeleton. We believe it facilitates negative curvature necessary to generate these protrusions. I have provided coarse-grained molecular dynamics simulations, which, when paired with a mesoscale model implemented by my lab-mate (Ramakrishnan Natesan), predicts the molecular basis for the requisite membrane deformations. Observations of the perturbed undulations produced by exo70 inspired the curvature focusing story above. This paper is listed with my other publications.

divalent cations modulate phosphoinositide signaling
Most of my work employs coarse-grained simulation, however atomistic resolution is often required to understand lipid chemistry. In order to adequately characterize protein-lipid binding, it is necessary to understand the electrostatic interactions between highly-charged lipids with both their protein binding partners and the tightly-regulated pools of divalent cations present in the cell. We investigate these interactions using atomistic simulation in order to better understand how key signaling lipids are organized on the plasma membrane. I have contributed simulations to understand actin nucleation (2019, 2020), and divalent cation binding under various conditions,

modeling receptor flexure to understand nanocarrier binding
The extracellular space has a rich composition which provides both a substrate for the cell as well as a medium by which they interact. Scientists and engineers who wish to design targeted therapeutics require a detailed understanding of the mechanics of protein receptors which can tether cells to extracellular cargo. At the beginning of graduate school, I contributed coarse-grained simulations of intracellular adhesion molecule-1 (ICAM-1) to a multiscale model for nanocarrier binding to endothelial cells. Such multiscale models are necessary to optimize the nanocarrier design so that it targets particular cell populations. My original contribution is listed under publications, however extensions to this work are ongoing.
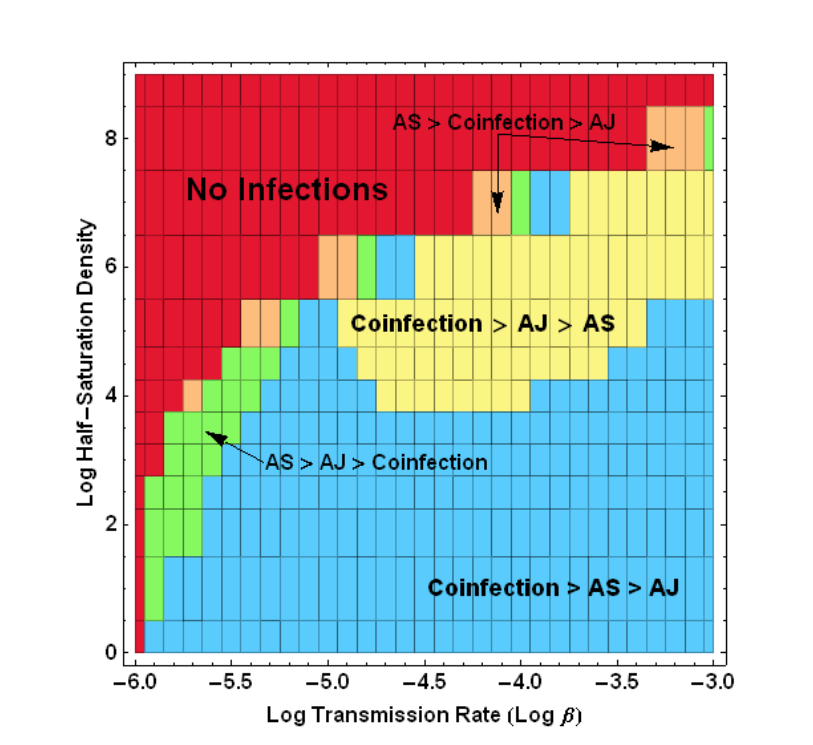
Phase diagram of diseased populations inferred from within-host virus measurements. We find a crescent-shaped boundary characterizes the diseased state, and in some places, coinfections are not dominant.
modeling the spread of malaria
As an undergraduate, I developed a model for understanding the spread of malaria in human populations. Malaria parasites reproduce in the bloodstream and are transmitted between humans by mosquitoes. The number of parasites in the blood can therefore affect the transmissibility of the disease and its persistence in the human population. In my undergraduate thesis, I present a model which captures the population dynamics of wild-type and drug-resistant malaria according to the course of the disease in mouse experiments provided by collaborators. This model can be used to predict the emergence of drug-resistant infections. Similar methods may be used to design optimum treatment strategies for delaying the emergence of drug-resistant strains. For more details, download my undergraduate thesis.
 Ryan Bradley, Ph.D.
Ryan Bradley, Ph.D.